Beyond “Junk DNA”: Non-coding RNAs as a Regulatory Layer
Large portions of the human genome once dismissed as “junk DNA” are now recognized as transcriptionally active. High-throughput sequencing has revealed pervasive transcription and a diverse repertoire of non-coding RNAs (ncRNAs) with regulatory potential.
This paradigm shift does not imply universal functionality. While some ncRNAs show tightly regulated, cell-type-specific or stress-responsive expression, others likely reflect transcriptional noise or RNA processing byproducts. Discriminating functional ncRNAs from incidental transcripts is essential, particularly when proposing mechanistic, diagnostic, or therapeutic roles.
Length-Based Classification: Practical but Incomplete
ncRNAs are commonly categorized by length into small ncRNAs (<200 nt) and long ncRNAs (>200 nt), a framework that remains useful for experimental and computational workflows.
However, circular RNAs (circRNAs) challenge this classification. Despite often exceeding 200 nt, circRNAs are defined by covalent circularization rather than size. This topology confers distinct biochemical properties, including resistance to exonucleases, separating circRNAs functionally from linear long ncRNAs.
Exosomes as Extracellular RNA Carriers
Exosomes are endosome-derived extracellular vesicles (30–150 nm) that transport RNA protected within a lipid bilayer, conferring stability in RNase-rich biofluids.
Importantly, exosomes are not the sole carriers of extracellular RNA. Many circulating RNAs are bound to ribonucleoprotein complexes, such as AGO2-associated miRNAs or HDL-bound RNAs. Moreover, while exosome-mediated RNA transfer can be biologically relevant, functional effects in recipient cells are often context-dependent and quantitatively limited.
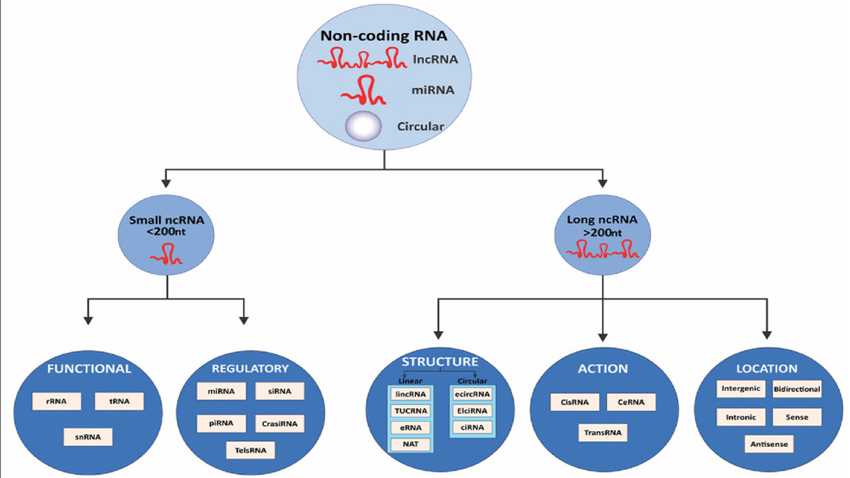
Classification of noncoding RNAs (ncRNAs)
https://www.researchgate.net/figure/Classification-of-noncoding-RNAs-ncRNAs-Noncoding-RNAs-are-classified-into-small_fig1_339894753
Diversity of ncRNAs in Exosomes
Most major ncRNA classes have been detected in exosomes, but their abundance and enrichment vary widely. RNA profiles are influenced by cell type, physiological or pathological state, and technical factors, particularly vesicle isolation and sequencing methods.
Detection alone does not imply function. Some RNA species—such as snRNAs or snoRNAs—may reflect selective export in specific contexts, but in others may indicate nuclear RNA turnover, stress responses, or dysregulated transcription.
MicroRNAs (miRNAs)
miRNAs are the most consistently enriched and extensively studied exosomal RNAs. Their sorting is mediated by RNA-binding proteins such as hnRNPA2B1, YBX1, and SYNCRIP, often via defined sequence motifs (“EXOmotifs”).
Although exosomal miRNAs can modulate gene expression in recipient cells, effect sizes depend on miRNA copy number, vesicle uptake, and RISC loading. Reports of strong or rapid regulatory effects should therefore be interpreted cautiously.
Long Non-coding RNAs (lncRNAs)
Tumor-derived exosomes are often enriched in specific lncRNAs, including HOTAIR, MALAT1, and H19. Experimental studies support roles in fibroblast reprogramming, angiogenesis, and immune modulation.
However, the strength of evidence varies substantially. Many reported associations remain correlative, highlighting the need for rigorous functional validation.
Circular RNAs (circRNAs)
circRNAs are highly stable in extracellular environments, contributing to their enrichment in exosomes and utility as biomarkers. Active export may also serve as a cellular clearance mechanism.
Although frequently described as miRNA sponges, this function is often overstated and context-dependent, with stoichiometry frequently insufficient for meaningful effects. Alternative roles—including protein binding, transcriptional regulation, and translation into micropeptides—are increasingly recognized.
tRNA-Derived Fragments (tRFs)
tRNA-derived fragments and stress-induced tiRNAs are among the most abundant small RNAs in exosomes across biofluids. These molecules are not random degradation products but participate in translational repression, stress signaling, metabolic regulation, and immune responses.
Their exosomal enrichment likely reflects both regulated export and increased tRNA cleavage under cellular stress.
Additional ncRNA Species
Exosomes also contain less frequently discussed ncRNAs, including Y RNAs and their fragments, vault RNAs, and repeat-derived RNAs. These species contribute to extracellular RNA signatures and may hold diagnostic or functional relevance.
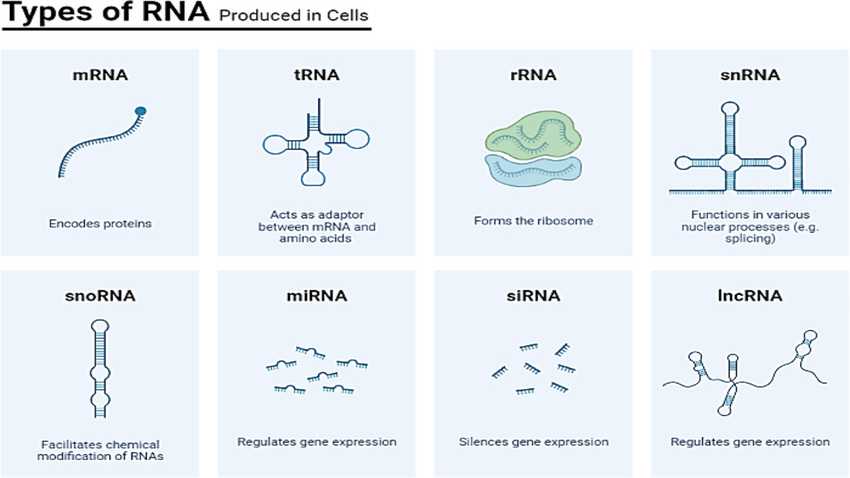
Diversity of types of RNA produced in cells
https://www.researchgate.net/publication/377437774_Integrative_role_of_small_non-coding_RNAs_in_viral_immune_response_a_systematic_review
Exosomal ncRNAs in Liquid Biopsy
Exosomal ncRNAs are actively investigated as biomarkers for cancer, neurodegenerative disorders, and other diseases. Their stability and disease-associated expression patterns make them attractive for liquid biopsy applications.
Clinical translation remains limited by challenges including variable isolation methods, contamination by non-vesicular RNA, and substantial inter-individual variability. Only a small number of assays have achieved robust clinical validation.
A Balanced Perspective
Exosomes carry a diverse but non-random repertoire of ncRNAs—miRNAs, lncRNAs, circRNAs, tRNA-derived fragments, and other small RNAs—reflecting both selective sorting mechanisms and the state of the originating cell. This complexity underpins their promise for studying intercellular communication and biomarker discovery, while demanding methodological rigor and restraint in functional interpretation.
 ️
️